Oncology
Part-per-million accuracy
with low costs
The Ultima Advantage
A comprehensive picture of the tumor signature through whole genome sequencing of the tumor and germline provides a rich dataset to inform follow-on monitoring in plasma. Achieve this with cost-effective sequencing on the UG 100.
Sequence whole genomes on UG 100
Reference: Zviran et al. Nat Med. 2020 Jul;26(7):114-1124
Achieve low LoD cost-effectively:
The combination of high throughput, low costs and extreme accuracy for SNVs enables part-per-million accuracy without extreme sequencing depth through over-sequencing.
Opportunity for more frequent monitoring:
With lower sequencing costs, it becomes more practical to support frequent cancer recurrence monitoring strategies in hopes to detect cancer recurrence earlier.
Learn about ppmSeq™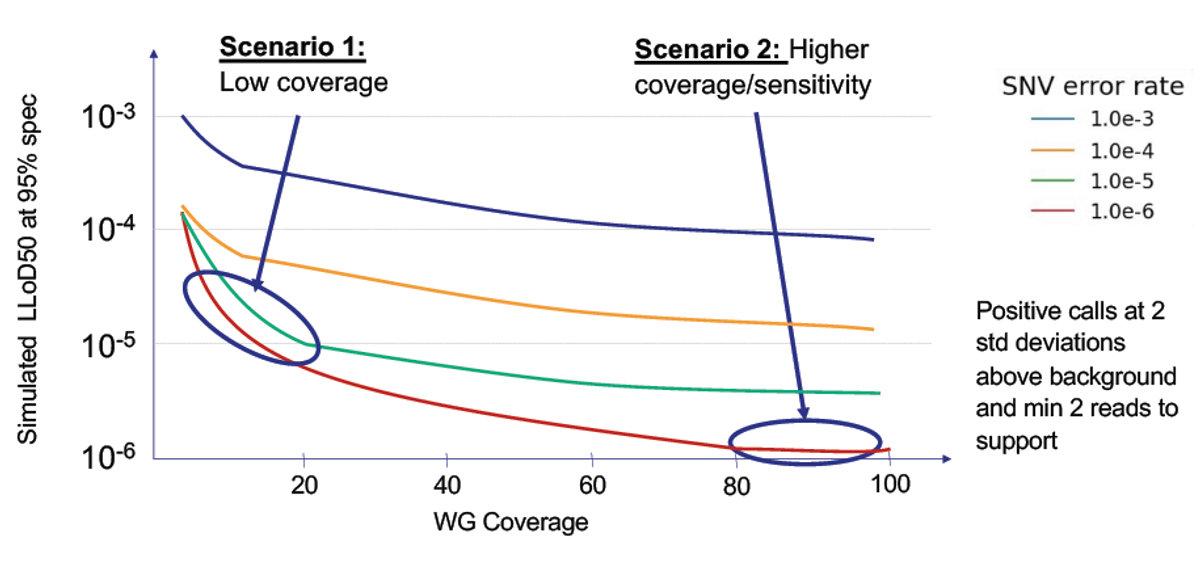
ppmSeq™ for extreme SNV accuracy
The combination of Ultima’s flow-based sequencing chemistry and emulsion-based clonal amplification technologies enables a ground-breaking new method: paired plus minus sequencing, or ppmSeq™. By sequencing both strands of a captured DNA molecule, ppmSeq reduces errors to one part-per-million or better, empowering scientists to tackle the most challenging sequencing applications.
The frequency of these rare events is not only lower than traditional sequencing error rates, but also lower than the error rate of common sample artifacts.
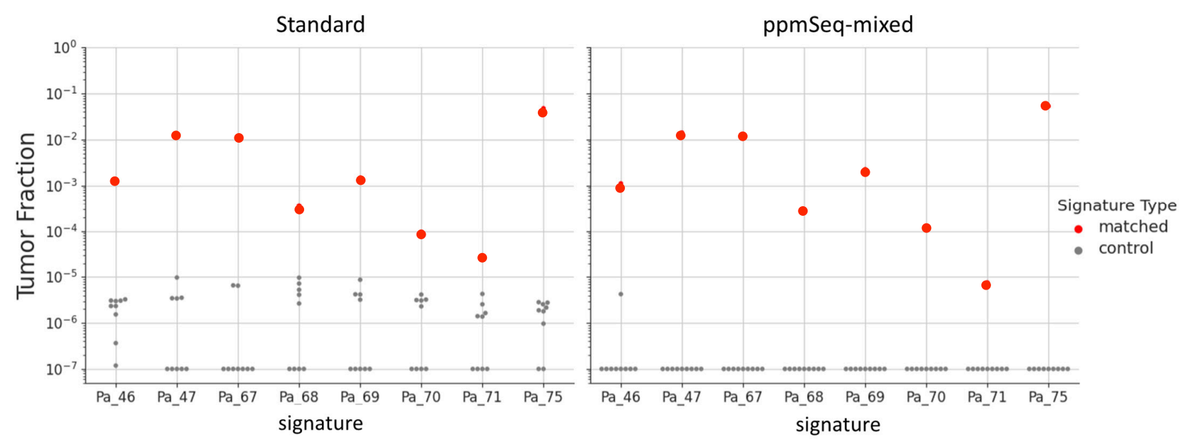
Estimated tumor fraction in matched and control cfDNA samples, for standard cfDNA prep (left) and ppmSeq-mixed (right). Each column corresponds to one patient SNV profile, with one matched cfDNA sample and 9 negative control samples. Included are reads that exceed the quality thresholds of SNVQ60 for Standard, and SNVQ70 for ppmSeq-mixed.
Unique chemistry for SNVs
Our unique flow-based chemistry inherently provides advantages for calling SNVs. Our chemistry asks how many nucleotides are added, but the base identity is not a question. Incorporating one type of nucleotide per flow enables an extremely low base substitution error rate.

SNVQ is the single nucleotide variant quality (SNVQ) score, which is different from a standard base quality score. The SNVQ score represents the error probability of the specific base substitution (e.g., A>G) rather than the aggregate error probability of any substitution (e.g., A>C/G/T).
MRD with the UG 100™ Solaris Free workflow
The UG 100 Solaris™ Free workflow offers PCR-free library prep compatible with leading library prep kits and accommodates a range of sample types and input amounts.
Explore Solaris library prepView library compatibility guideHighlights
| PCR-Free Whole Genome Sequencing | PCR-Free liquid biopsy with ppmSeq | |
|---|---|---|
| Adapter names | xGen™ PCR-Free Adapters for Ultima Genomics® | UG ppmSeq™ Adapters |
| Purchase from | IDT | Ultima Genomics |
| Sample indexing | 192 sample indices | 96 sample indices |
| DNA type | Genomic DNA (gDNA) | Cell-free DNA (cfDNA) |
| DNA input range | 100 - 500 ng* | 1 - 20 ng* |
*DNA input amounts verified by Ultima Genomics.
Community testimonial
We are excited to introduce MRDVision as the first WGS-based MRD solution developed on Ultima's ppmSeq technology. MRDVision will be a game-changer in the field, providing groundbreaking sensitivity and genomic coverage.”
Jehee Suh | Chief Executive OfficerInocras, October 2024
Start saving now.
Run your next project on the UG 100.
Access our technology

Certified Service Providers
Access cost-effective and scalable sequencing through our growing network of global service providers vetted for technical expertise and adherence to best practices. Submit samples and start generating data on the UG 100 Sequencer now.
Sequence with a CSP todayTechnology Access Program
Our Technology Access Program offers accessible entry to the UG 100™ through our in-house applications lab. Sequence on our high-capacity fleet of UG 100 Sequencers and receive support at every step, from consultation through data analysis.
Discover our TAP offeringsBring UG 100 into your lab
Interested in exploring how your lab can benefit from high-throughput, cost-effective sequencing? Learn how our novel use of open wafers and built-in automation is continuously driving the cost of sequencing down to accelerate omics at scale.
Explore our technology